GRINs
GRIN1 and GRIN2 Genes Encoding NMDA Receptors
The NMDA receptors are tetrameric complexes of two GluN1 proteins (encoded by GRIN1) and two GluN2 subunits (encoded by GRIN2), and require simultaneous binding of the co-agonists glycine (to GluN1) and glutamate (to GluN2) for activation. The NMDA receptors play a special role in synaptic transmission. These receptors mediate a strong Ca2+-influx during synaptic activation due to high Ca2+ permeability and prolonged response time course. The resulting increase in intracellular Ca2+ can trigger multiple downstream signaling events in the postsynaptic neuron that can produce both short-term and long-term changes in synaptic efficacy or morphology, known as synaptic plasticity. Thus, the frequency and duration of synaptic NMDA receptor activation can result in either potentiation or depression of synaptic efficacy, which is considered a cellular correlate of memory and learning.
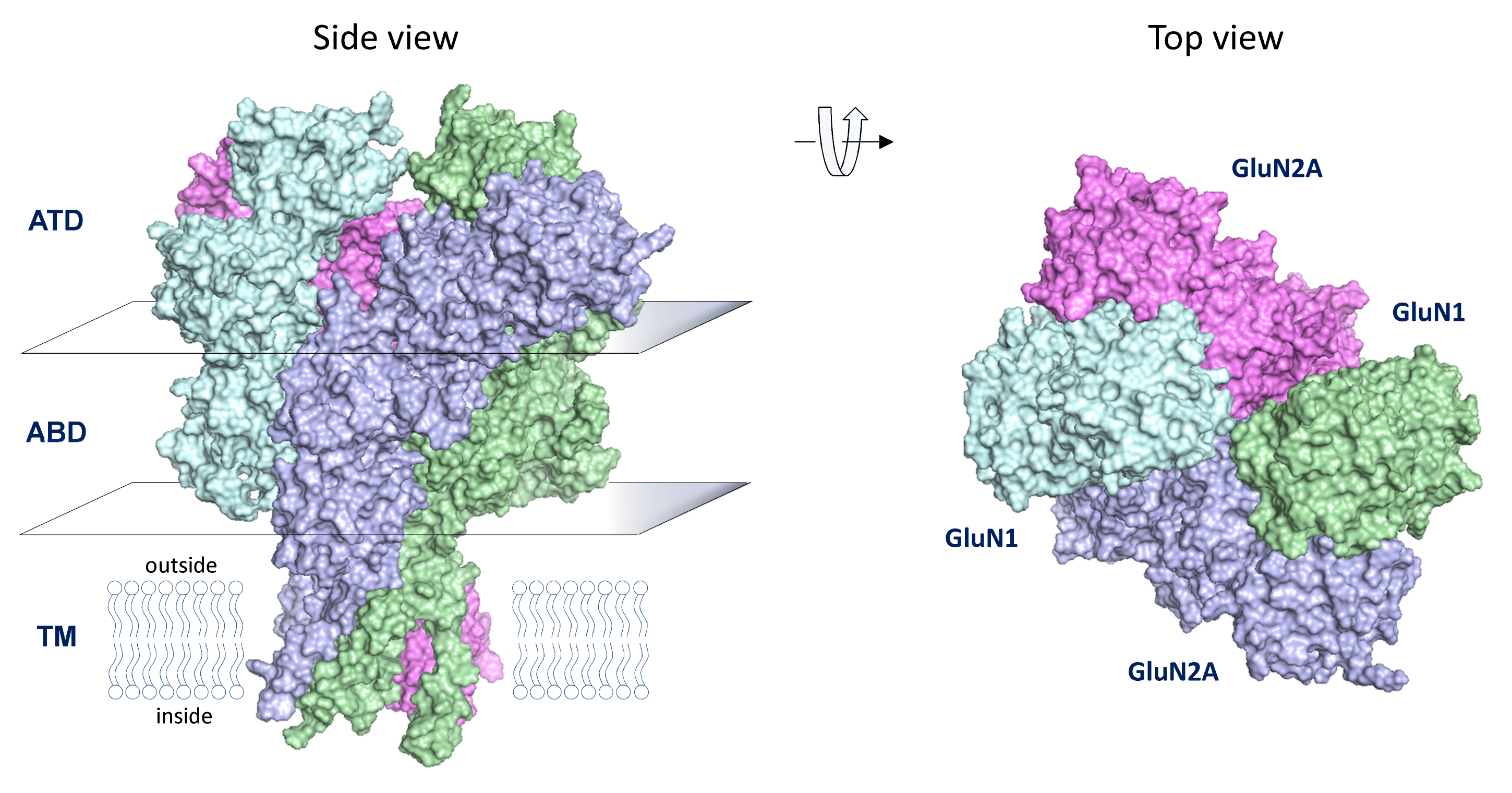
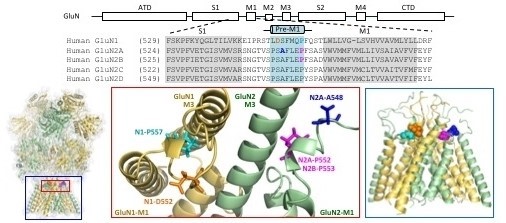
GluN1/GluN2A NMDA Receptor Structure based on Karakas and Furukawa, Science 344:992 (2014) and Lee et al. Nature 511:191 (2014) from P. Burger, H. Yuan, and S. Traynelis as reported in Chen et al. Mol. Pharm.91: 317 (2017).
GRIN3 genes encoding glycine binding GluN3 subunits
GluN3A and GluN3B proteins are encoded by GRIN3A and GRIN3B, and share sequence similarity to both GluN1 and GluN2 subunits. The GluN3 subunits bind glycine and D-serine, but the functional properties and physiological roles of GluN3-containing NMDA receptors remain elusive. Biochemical analysis of the protein composition of native glutamate receptors suggest that NMDA receptors may be assembled from a combination of GluN1, GluN2, and GluN3 subunits in the central nervous system. However, we still do not know the subunit stoichiometry of the potential triheteromeric GluN3-containing NMDA receptors, as it is unknown whether GluN3 replaces only GluN1 or GluN2, or alternatively, both GluN1 and GluN2 in these receptors. Despite this gap in our understanding, there is data that suggests that the inclusion of GluN3 in the NMDA receptor complex reduces Mg2+-block and Ca2+-permeability as well as response amplitudes.
The GluN3 subunits can also assemble with GluN1 in heterologous expression systems to form functional receptors that comprise two GluN1 and two GluN3 subunits. However, the assembly of these receptors in the CNS is not firmly established. GluN1/GluN3 receptors have been termed “excitatory glycine receptors”, since they can be activated by glycine alone without the requirement of glutamate binding, and mediate an inward current when a cation-selective pore is opened following glycine binding. In recombinant systems, the GluN1/GluN3 receptors show low Ca2+-permeability and are insensitive to extracellular Mg2+, two features that are distinct from classical GluN1/GluN2 NMDA receptors.
GRIAs
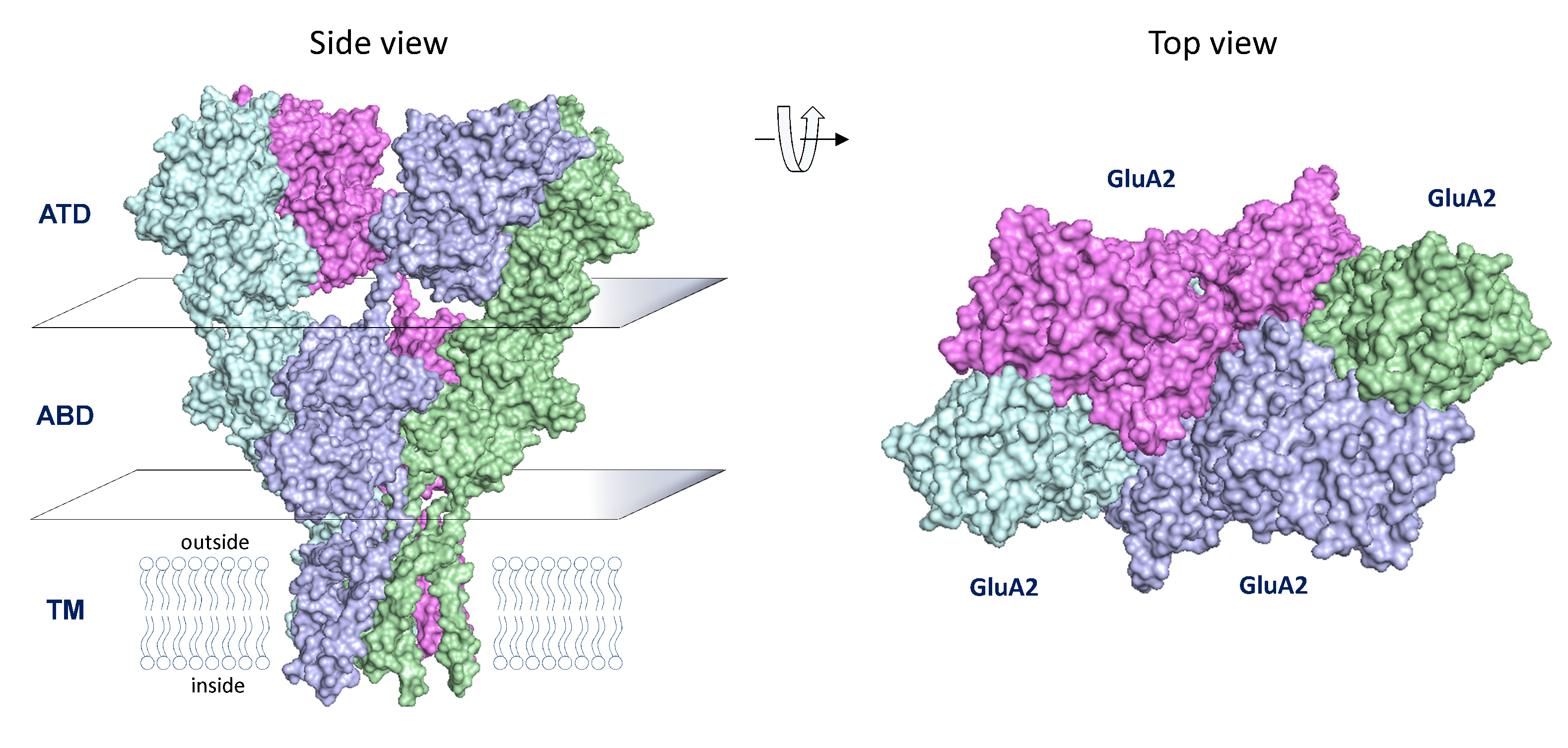
GRIA Genes Encoding AMPA Receptors
AMPA receptors are tetrameric assemblies of GRIA1-4 gene product and mediate a fast synaptic current. Glutamate can bind to each subunit, and triggers opening of an incrementally larger pore as more subunits bind glutamate. The AMPA receptors rapidly close
GRIKs
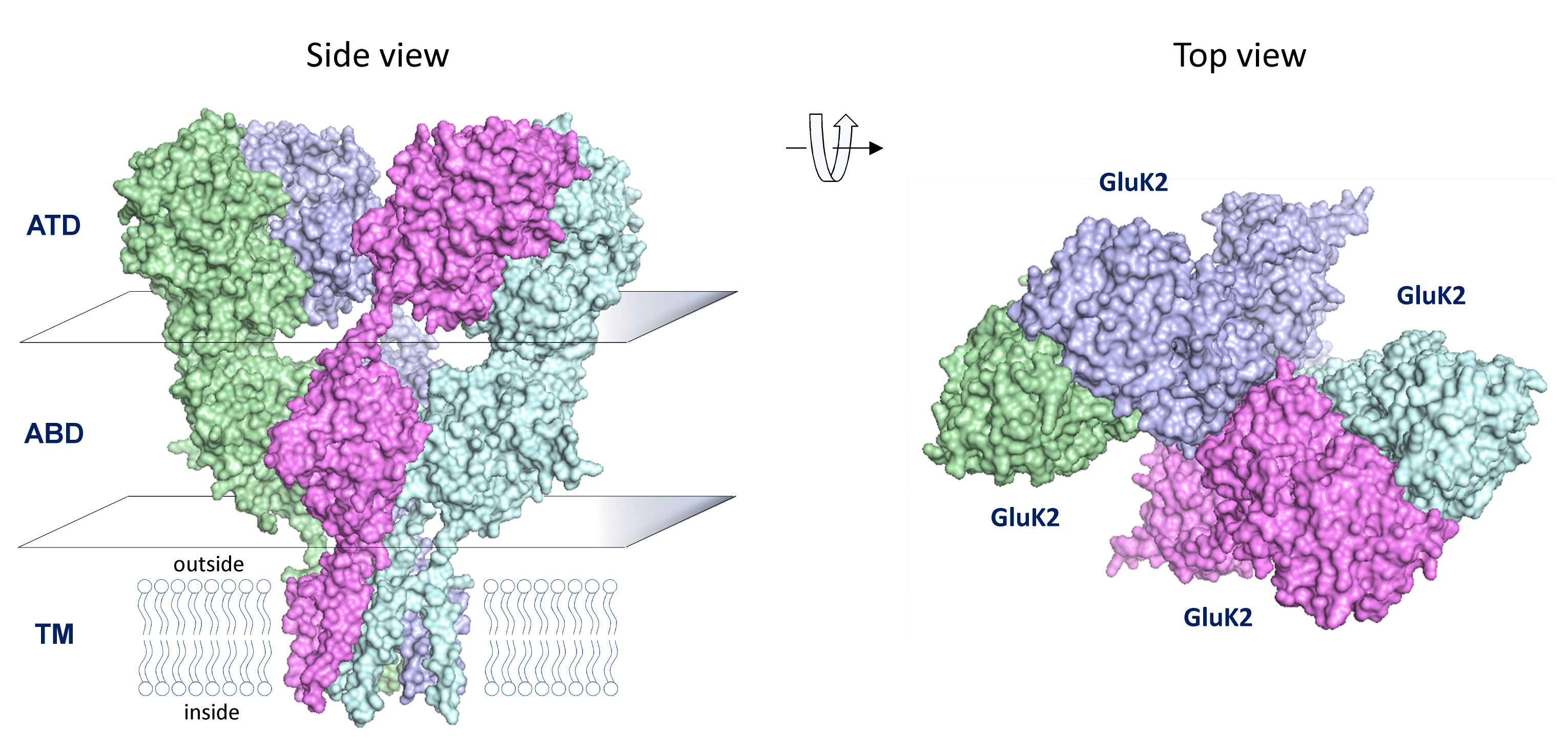
GRIK Genes Encoding Kainate Receptors
Kainate receptors are tetrameric assemblies of GRIK1-5 gene products. Activation of postsynaptic kainate receptors typically result in EPSCs with a slower time course than AMPA receptors and a comparable, but generally faster time course than NMDA receptors. Kainate receptors undergo desensitization in the presence of glutamate, which is modified by the Neto family of accessory subunits. Kainate receptors undergo variable RNA editing, which can alter their permeability to Ca2+.
GRIDs
GRID genes encoding GluD subunits
GluD1 and GluD2 are two proteins with significant sequence similarity to other members of the glutamate receptors family, and are encoded by GRID1 and GRID2. These receptors, known as δ (delta) receptors, are poorly understood. It is unclear whether they form functional ion channels in the brain, yet they appear to have important roles in neuronal development. These transmembrane proteins may form binding partners for presynaptic proteins to play a role in synapse organization and some forms of synaptic plasticity.