Mission
Center for Functional Evaluation of Rare Variants (CFERV): Background and Approach
The Problem: A large number of early life disorders, including epilepsy and developmental delay, involve genetic variations, most often de novo mutations. These devastating conditions can result in refractory seizures, developmental delay, and life altering medical issues that create an enormous burden on families.
Recent Progress: Stunning advances in DNA analysis have increasingly enabled definitive diagnoses to be made, reducing unnecessary testing, bringing clarity to the family, and focusing basic and clinical research resources. These diagnoses have yielded clinical insight, for example showing that the majority of epilepsies that start prior to one year of life (e.g. infantile epileptic encephalopathies) occur due to de novo genetic errors.
Current Knowledge Gap: A large and expanding gulf has developed between genetic information describing rare variants and de novo mutations in patients and our understanding of how these genetic variants affect the function of the proteins they encode. The lack of functional understanding blunts the promise that genetic analysis holds for improved diagnosis and effective treatment. Furthermore, it prevents translation of genetic information into a better understanding of the basis for disease. This imbalance between genetic and functional information exists because (1) the rate at which sequencing can be accomplished far exceeds the rate at which functional data can be collected, (2) basic functional evaluation of expressed gene variants, proteins, is expensive and requires unique protein-specific assays, and (3) there are few coordinated and systematic efforts to address functional consequences of genetic variation in a manner that is nationally accessible.
Our Approach: Strong genetic, statistical, and biological data support the idea that ion channel variants contribute to and give rise to disease, particularly neurological disease. CFERV was created with support from the National Institute of Neurological Disorders and Stroke (NINDS), Emory University, and generous philanthropic donors to partner basic scientists, geneticists, and clinicians to experimentally assess the functional consequences of genetic variants identified in clinics across the country. This effort has been initiated around a gene family for which we have a large set of mutations, the glutamate receptor genes GRIN, GRIA, GRIK, and GRID. CFERV serves as a national clearing house for mutations in this gene family, with the goal of providing experimental data on the functional consequences of each mutation.
CFERV has clinical partners in all 50 states, as well as multiple international partners. CFERV’s activities will catalyze more accurate diagnoses, advance an understanding of disease mechanism, and suggest therapeutic opportunities for applied precision medicine.
Future Directions: CFERV seeks to broaden this effort to include many gene families.
Scope
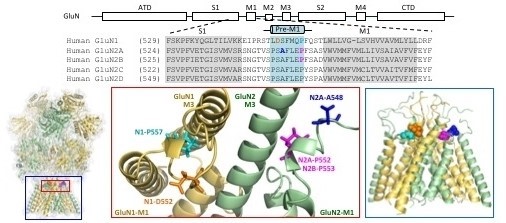
CFERV evaluates whether glutamate receptor function is altered by rare genetic variants within the associated family of human genes, GRIN (7), GRIA (4), GRIK (5) and GRID (2). These genes encode ligand-gated ion channels known as NMDA, AMPA, KA, and delta receptors, which play essential roles in brain function. Thus, mutations can play a causative role in neurological disorders and disease.
Rare variants in one or more of these genes (see Table) have been identified in patients with epilepsy syndromes, intellectual disability, developmental delay, autism spectrum disorder, schizophrenia, attention deficit hyperactivity disorder, hypotonia, cerebral visual impairment, and other conditions.
CFERV accepts requests to evaluate patient-associated rare variants that include missense, nonsense, frameshift, or internal protein coding deletions. All requests are carefully considered and when selected for study, the mutation is introduced into a human cDNA encoding the target subunit and receptor function is then evaluated in a series of electrophysiology and biochemistry studies.
|
Genes |
Chromosomes |
Protein Subunits |
Endpoints measured |
|
GRIN1, GRIN2A, GRIN2B, GRIN2C, GRIN2D, GRIN3A, GRIN3B |
9q34.3, 16p13.2, 12p12, 17q25, 19q13.1-qter, 9q31.1, 19p13.3 |
GluN1, GluN2A, GluN2B, GluN2C, GluN2D, GluN3A, GluN3B |
Agonist glutamate potency (EC50) |
|
GRIA1, GRIA2, GRIA3, GRIA4 |
5q31.1, 4q32-q33, Xq25-q26, 11q22 |
GluA1, GluA2, GluA3, GluA4 |
|
|
GRIK1, GRIK2, GRIK3, GRIK4, GRIK5 |
21q22.11, 6q16.3-q21, 1p34-p33, 11q22.3, 19q13.2 |
GluK1, GluK2, GluK3, GluK4, GluK5 |
|
|
GRID1, GRID2 |
10q22, 4q22 |
GluD1, GluD2 |