Large-scale profiling methods have uncovered numerous gene and protein expression changes that correlate with tumorigenesis. However, determining the relevance of these expression changes and which biochemical pathways they affect has been hindered by our incomplete understanding of the proteome and its myriad functions and modes of regulation. Activity-based profiling platforms enable both the discovery of cancer-relevant enzymes and selective pharmacological probes to perturb and characterize these proteins in tumor cells. When integrated with other large-scale profiling methods, activity-based proteomics can provide insight into the metabolic and signaling pathways that support cancer pathogenesis and illuminate new strategies for disease diagnosis and treatment. Overall goal of our lab is to understand how alterations in the function of proteins contribute to human disease and to identify individual proteins and cognate biochemical pathways that can be pharmacologically targeted for human disease. Therefore our laboratory is mainly focused on developing of novel chemical probes for profiling poorly characterized protein activities and drugs for prevention, diagnosis, treatment and cure of diseases like cancer and diabetes.
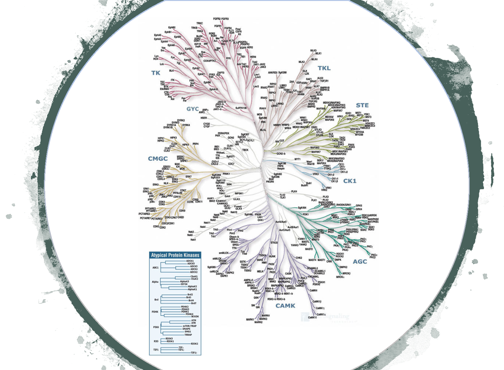
1. Development of chemical proteomic probes for profiling kinase activity
Protein kinases are a large family of proteins that serve key roles in nearly all aspects of cellular function. When deregulated, these enzymes have been shown to contribute to human diseases ranging from diabetes to cancer. Despite intensive research efforts, the physiological function, and specifically the context-dependent activity states, of the majority of protein kinases remain unknown due to a lack of technologies to study kinase function within cells, tissues and whole organisms. We aim to develop selective chemical proteomic probes that will report on the activity of all kinases in a cell or tissue in parallel.
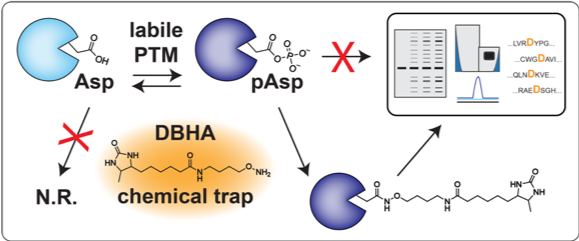
2. Identification of novel bacterial virulence factors by activity-based protein profiling
The virulence factors of bacterial pathogens and the regulatory systems that monitor their expression are predominantly attractive for therapeutic intervention. Recently, Two-component signaling systems (TCSs) have emerged as potential targets for antibacterial drug design for a number of reasons. As a common practice, conventional antibiotics target specific bacterial proteins that perform essential functions in the pathogen; however, a drug that targets TCSs could be highly effective because it impairs upstream regulatory functions related to the pathogen’s physiology instead of affecting only specific downstream functions. Thus, the use of TCSs for drug development provides an alternative strategy for combating microbial infections, including those caused by pathogens resistant to currently available antibiotics. We will use the Desthiobiotin-containing O-alky hydroxylamine (DBHA) probe to identify novel bacterial virulence factors that are active in animals infected with multidrug-resistant (MDR) pathogens.
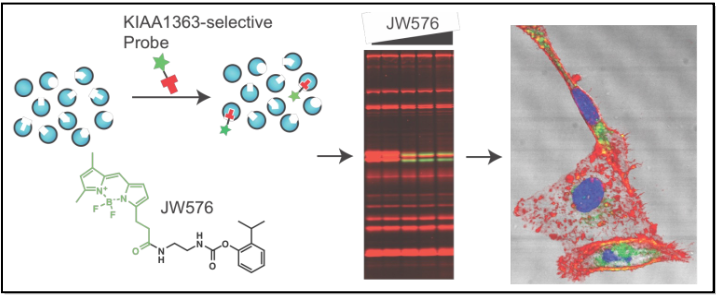
3. Developing precision imaging probes for metastatic cancer
Historically, tumor imaging has primarily relied on the use of radiotracers, the most popular of which is 18fluorodeoxyglucose (FDG), for positron-emission tomography (PET) imaging of tumor cells in vivo. Two major limitations of most of these agents, including FDG, are: poor specificity for tumor tissue that results from background activity in normal tissues; variable residence times bound to the target of interest, which in many cases limits clinical utility. The successes and limitations of these clinical diagnostics validate the need to identify new biologic targets associated with cancer as well as molecules capable of specifically targeting them for diagnostic imaging purposes. We will develop protein-specific, rather than class-specific, imaging probes that covalently modify intracellular targets of interest with high potency and specificity.

